I have had 4 mains sequences with overlapping and I have published 182 papers.![]()
Here are only the milestones steps.
Immunochemistry of Staphylococcus aureus

• The isolation and characterization of various protein (Flandrois, Fleurette, & Modjadedy, 1975; Grov, Flandrois, Fleurette, & Oeding, 1978) or polysaccharides (Ndulue & Flandrois, 1983) was the following of my PhD Thesis. I have also described the affinity of the cells to human proteins (Carret et al., 1985).
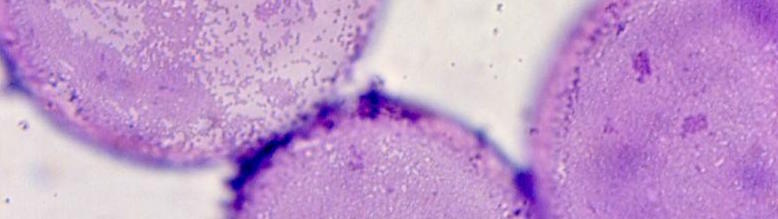
• A side effect of this research has been a world-wide used affinity reagent (Carret, Flandrois, Bismuth, & Saulnier, 1982) (“StaphySlide”) that remains, in a revised version, the basis of most of the immediate identification reagents for Staphylococcus aureus (photography above: StaphySlide, the red-cells covered with fibrinogen and S.aureus adhering).
Modelling of bacteria growth
Applications to antibiotics sensitivity and food safety
• Modelling of bacteria growth was made possible by the early development of informatics. My team was involved in the mathematical approach and optimal production of biological datas. we have demonstrate the change of growth model occuring when bacteria are exposed to the drugs and described the mathematical law linking growth rate and other parameters and antibiotics concentration. This was done both for sub-inhibitory concentrations (Comby, Flandrois, Carret, & Pichat, 1988) and lethal concentrations (Guérillot, Carret, & Flandrois, 1993). A major paper describe the interpretation of the antibiotic effect in term of maintenance energy for the cell (Lobry, Carret, & Flandrois, 1992).
• The applications were not possible in real life due to the need of a dedicated high throughput performance photometer. We described also how photometry combined with mathematical modelling is able to study the swelling of the micro-organisms during the lag phase. The possibility to use our algorithms is now studied by research teams using the new “digital microbiology” concept.
• Aside this extensive model approach, I have developed the firrst “expert systems” to analyse and automatically correct the result of antibiotics susceptibility testing (Comby, Flandrois, & Pave, 1988). These “partially intelligent” programs are now included in each antibiotic susceptibility systems and derived from this original work.
• I oriented in parallel the team on the basic physiology of bacteria and we described then the biological-mathematical laws linking growth rate and latency to pH and temperature (Rosso, Lobry, Bajard, & Flandrois, 1995). More, we have discovered a fundamental law in the bacteria linking the optimum, minimum and maximum temperature of growth (Rosso, Lobry, & Flandrois, 1993). A lot of work was also done to model interactions between bacteria (especially those found in food, pathogens vs non pathogens) (Cornu, Kalmokoff, & Flandrois, 2002).
• There results and more works (Bréand, Flandrois, Rosso, Tomassone, & Fardel, 1997) were applied to the prediction of growth of bacteria in food in cooperation with Danone. The prediction of growth of pathogens in food is good at months level (up to 3 months). This “predictive microbiology” is now widely used in agro-food industry.
• Predictive microbiology is a part of the risk-analysis process in food, environment... (figure : simulation of the growth of Listeria monocytogenes in food with variable temperatures)
New methods for bacteria detection/enumeration

• This topic has been important all over my work, partly due to my position as Head of a diagnostic laboratory.
• I have been involved in patents concerning biomolecular detection of pathogens, new detection methods in food microbiology and use of digital microbiology in bacteria enumeration.
• This applied scientific research is my main preoccupation today:
- Detection of pathogens in food and water in a sealable container (Junillon 2014a,b,c, figure above corresponding)
- Detection of pathogens and contaminants in water in ESS conditions by molecular biology (Bechy 2015).
• What is important for me is that the two project have similar probability issues : the binomial law is to take into account and statistical signification of a given level of detection (especially 0) has to be accurately computed to compare the results. Earlier studies of my team on blood culture results were dealing with the same problem (Lamy 2002, Leyssene, 2012).
Bioinformatics applied to bacteria identification
This is the beginning of the on-going work
• The main result was the “BIBi” project (Devulder, Perriere, Baty, & Flandrois, 2003, Flandrois, Gouy and Perrière 2014) . I demonstrate that Bacteria Identification based on Bioinformatics (BIBi) was possible and easy to use in routine (S Mignard & Flandrois, 2006) if a convenient database was built. This program is available on the web and I recently release the mark 5 version using new algorithms to build the database and extract the pertinent information. Annually 50000 identi cations are done by the system and it is cited in 92 papers.
• As the Mycobacterium genus was the model genus that I use to develop BIBi (Devulder, Pérouse de Montclos, & Flandrois, 2005), I improve the phylogeny and taxinomic analysis of this genus by combining bioinformatics methods (Mignard & Flandrois, 2008, Guérin et al. 2014, Pin 2014). .