Phylogeny for the microbiologist
Being involved both in microbiology and bioinformatics I feel necessary to develop solutions to give a easy access to microbial phylogenetic reconstructions for non specialists
The "Phylogeny on the Fly" (PPF) project
"Phylogeny on the Fly" is a long-going project but we hope to publish our solution soon.
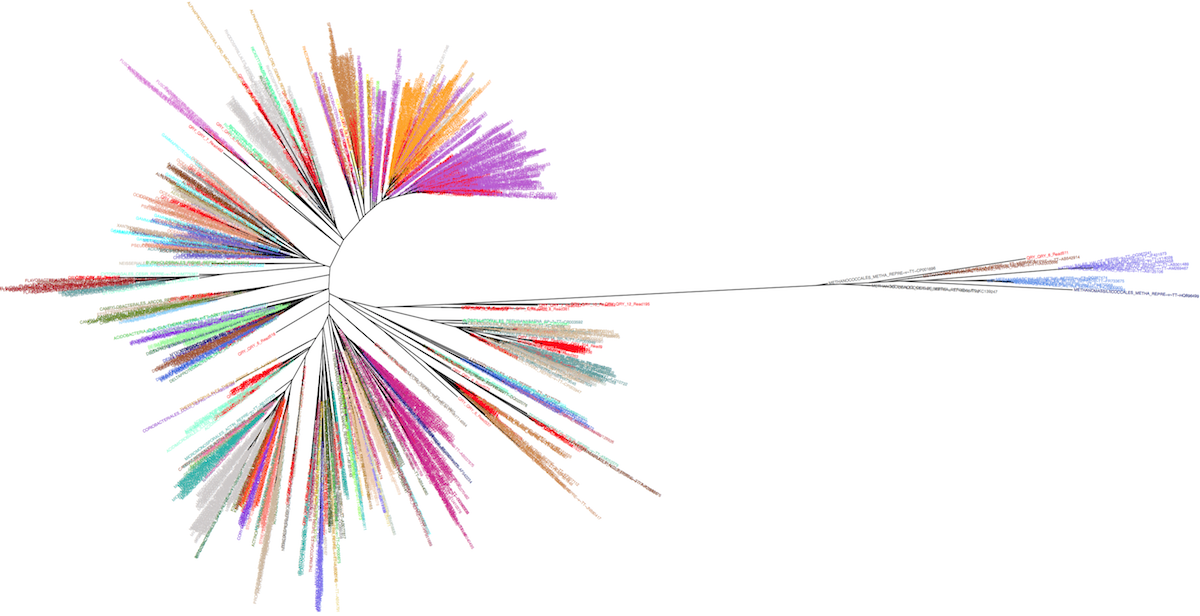
The idea is to take advantage of the leBIBI Russian doll databases to build phylogenies for a given level included in wider phylogenetic coverage without the overcrowding of non significant sequences. It is possible to extract high similarity sequences around a query at the species level or to extract sequences belonging to a given species or other levels (see the case-study of a laguna microbiome showing the position of the clusters nodes -in red- among Bacteria and Archaea).
Manolo Gouy and I have created an interactive webtool to represent the corresponding tree, enabling the visualisation of the desired taxonomic level information.
The "AmalPhy_lab" project

"AmalPhy" is not the acronym of "A Multi Agent Locus Phylogeny", even if this could have sense. The name was originaly a reference to the italian city and a joke based on a french approximate homophony. AmalPhy is not a real pipeline that take the output from a program to send to another one, it do this but with a dose of back-loops or recursivity and it construct a composite result using five different approach of the phylogenetic reconstruction for multi-locus phylogenies. Surprisingly I have been able to use many part if the PPF code and the last outputs may be viewed through PPF.
Amalphy V1 was written in 2007 for the Mycobacteria phylogeny paper. I have written the current version ("Amalphy_lab") in 2016 to enable a direct use (without human expertise or programming) of the outputs of RiboDB (figure, output of Actinobacteria phylogeny). The current version of AmalPhy is not available even if it is fully functionnal even for non-specialists, with no known bugs. This is because I need to interact with the users as no quality-control agent has been introduced up to now. I am open to cooperation with colleagues who want to use RiboDB (or other) to build multi-locus phylogenies.